EME: GDS
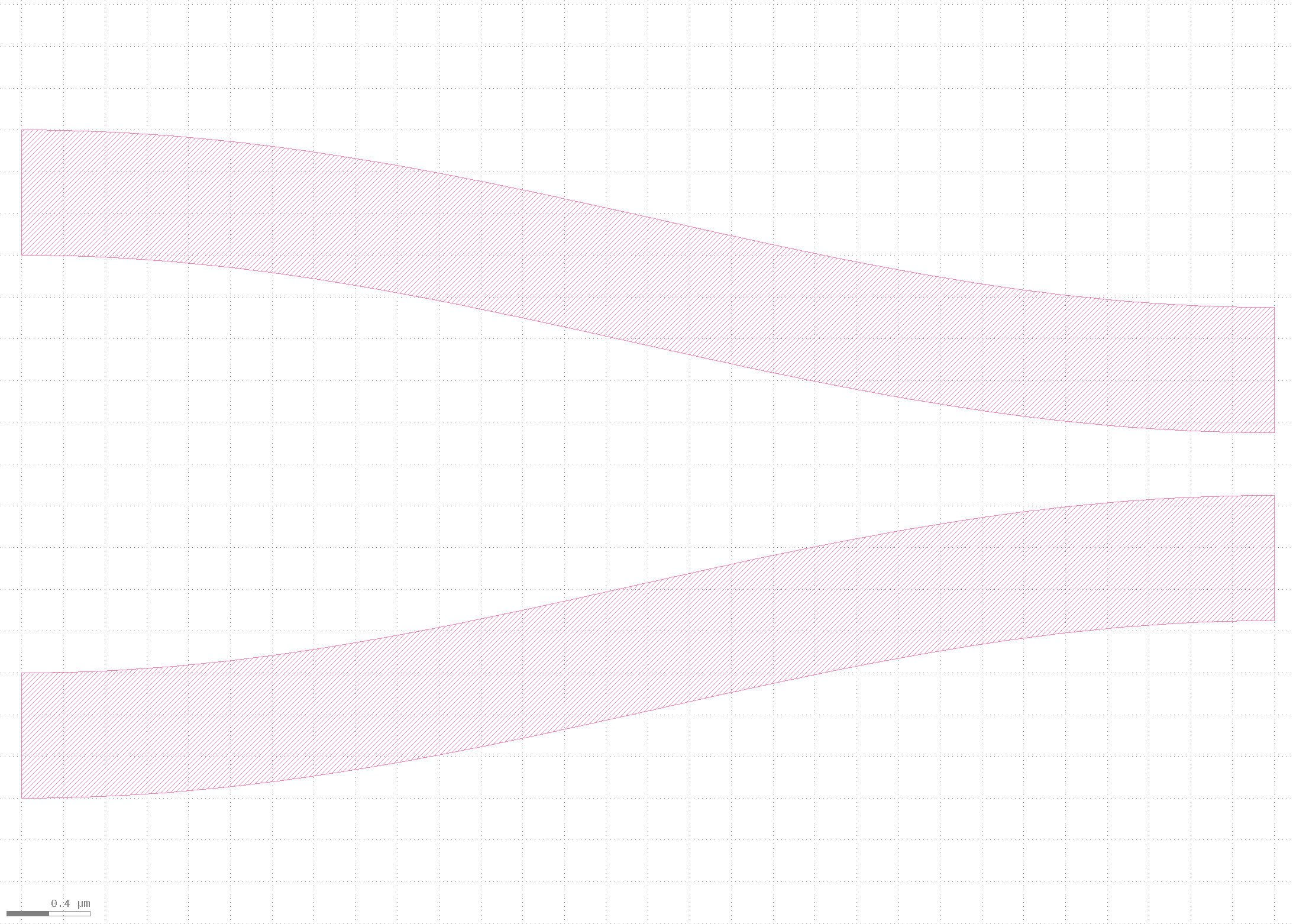
To simulate more complex structures, a GDS import function is used to support any geometry that can be created on a mask GDS file. This example shows how an s-bend structure can be created in a GDS file, imported to EMode, and simulated as part of a directional coupler.
This code example is licensed under the BSD 3-Clause License.
First, the GDS is created.
from phidl import CrossSection, Device
import phidl.path as pp
from phidl import quickplot as qp
## Parameters
width_wg = 0.6 # [um]
gap_input = 2.0 # [um]
gap_output = 0.3 # [um]
L_bend = 6.0 # [um]
## Create the first CrossSection
X1 = CrossSection()
X1.add(width = width_wg,
offset = -gap_input/2 - width_wg/2, layer = 1,
name = 'wg1', ports = ('in1', 'out1'))
X1.add(width = width_wg,
offset = gap_input/2 + width_wg/2, layer = 1,
name = 'wg2')
## Create the second CrossSection
X2 = CrossSection()
X2.add(width = width_wg,
offset = -gap_output/2 - width_wg/2, layer = 1,
name = 'wg1', ports = ('in2', 'out2'))
X2.add(width = width_wg,
offset = gap_output/2 + width_wg/2, layer = 1,
name = 'wg2')
## Transition from first to second CrossSection
Xtrans = pp.transition(cross_section1 = X1, cross_section2 = X2, width_type = 'sine')
P3 = pp.straight(length = L_bend)
WG_trans = P3.extrude(Xtrans)
## Place all CrossSection extrusions
D = Device()
wgt = D << WG_trans
## Quickplot to view
qp(D)
## Save gds file
D.write_gds('sbend.gds')
Next, the directional coupler device is created and simulated for a short coupler length. The sweep function is used to optimize the coupler length for 100 % coupling, and the propagation is re-plotted.
import emodeconnection as emc
from matplotlib import pyplot as plt
import numpy as np
## Set simulation parameters
wavelength = 1550 # [nm] wavelength
dx, dy = 20, 20 # [nm] resolution
h_core = 220 # [nm] waveguide core height
h_clad = 1200 # [nm] waveguide top and bottom clad
width_wg = 600 # [nm]
gap = 300 # [nm]
input_gap = 2000 # [nm]
window_height = h_core + h_clad*2
num_modes = 2 # [-] number of modes
BC = 'TE-v'
## Connect and initialize EMode
em = emc.EMode()
## Settings
em.settings(
wavelength = wavelength, x_resolution = dx, y_resolution = dy,
window_height = window_height, num_modes = num_modes, boundary_condition = BC, background_material = 'SiO2')
## Draw shapes
em.shape(name = 'BOX', material = 'SiO2',
height = h_clad)
em.shape(name = 'core', material = 'Si',
height = h_core, etch_depth = h_core)
## Create profiles
em.settings(window_width = 6200)
em.label_profile(name = 'slab')
em.settings(window_width = 2600)
em.shape(name = 'core', mask = width_wg)
em.label_profile(name = 'io_waveguide')
em.settings(window_width = 2600)
em.shape(name = 'core', mask = [width_wg, width_wg],
mask_offset = [-gap/2-width_wg/2, gap/2+width_wg/2])
em.label_profile(name = 'coupler')
## Draw EME sections
em.section(name = 'input', section_type = 'straight',
profile = 'io_waveguide', length = 1000,
offset = input_gap/2 + width_wg/2)
em.section(name = 'sbend', section_type = 'gds',
gds = 'sbend.gds', profile = 'slab',
shape_to_mask = 'core',
length = 10e3) # modify length of gds
em.section(name = 'coupler', section_type = 'straight',
profile = 'coupler', length = 30e3)
em.section(section_type = 'copy',
section_name = 'sbend', mirror = True)
em.section(name = 'output', section_type = 'straight',
profile = 'io_waveguide', length = 1000,
offset = -input_gap/2 - width_wg/2)
## Run EME and plot results
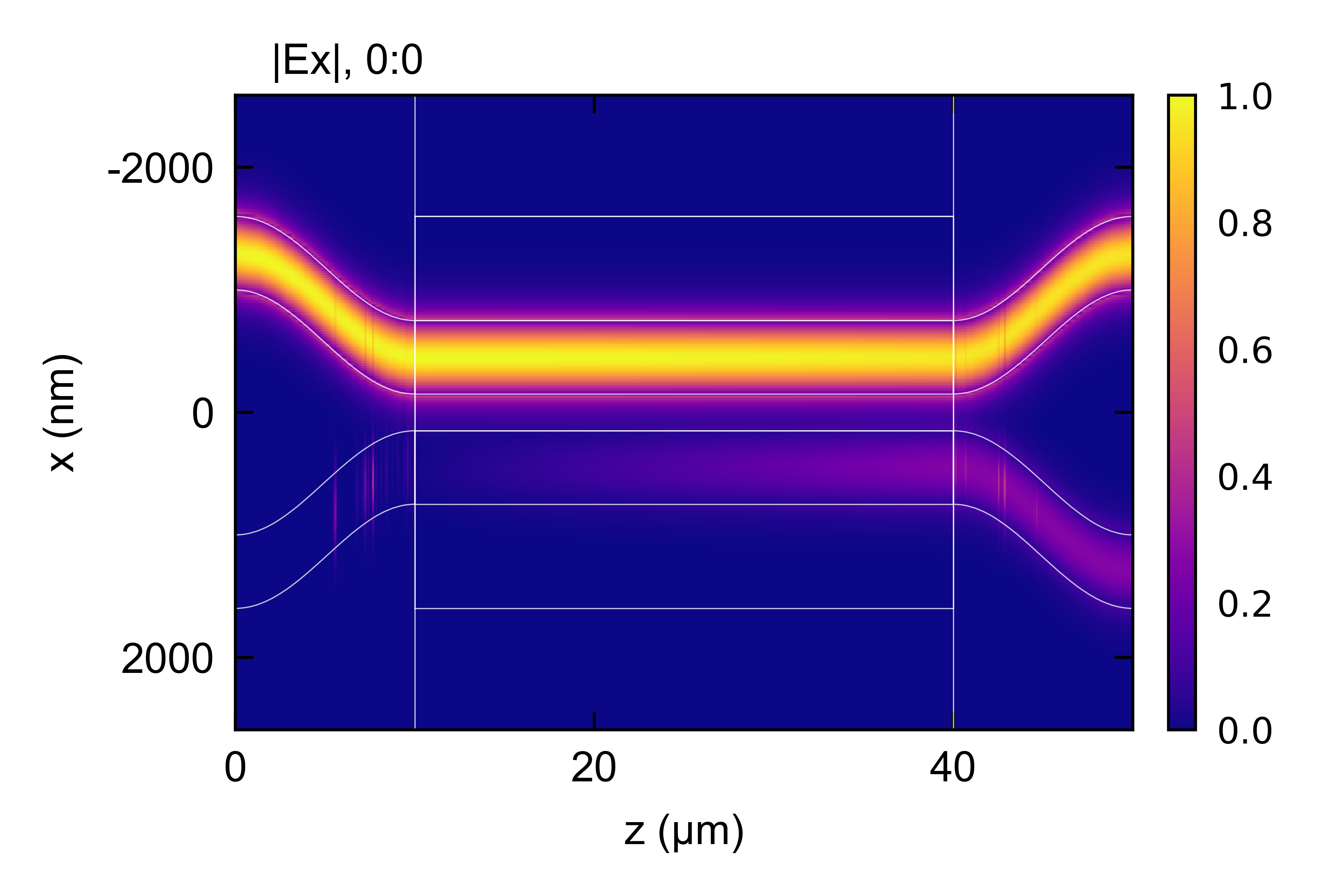
em.EME()
em.plot()
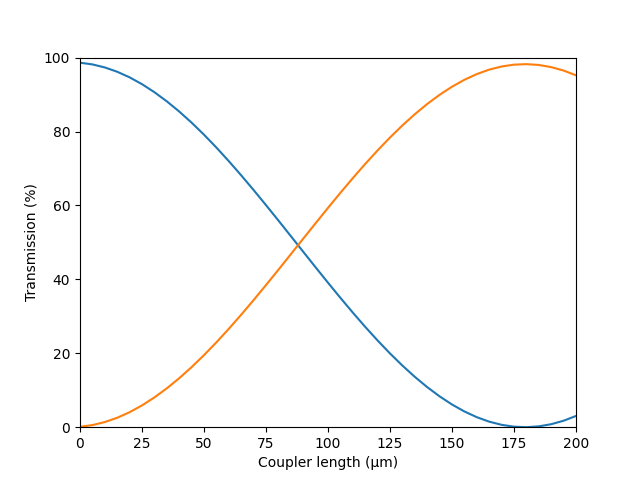
## Sweep the coupler length and plot results
data = em.sweep(key = 'section, coupler, length',
values = np.arange(0, 200.1e3, 10000),
result = ['S_matrix'])
S = data['S_matrix']
plt.plot(data['values']*1e-3, np.abs(S[:,1,0,0])**2*100)
plt.xlabel("Coupler length (\u03BCm)")
plt.ylabel("Transmission (%)")
plt.autoscale(enable = True, axis = 'x', tight = True)
plt.ylim([0,100])
plt.savefig('transmission_plot.png', dpi = 300, bbox_inches = 'tight')
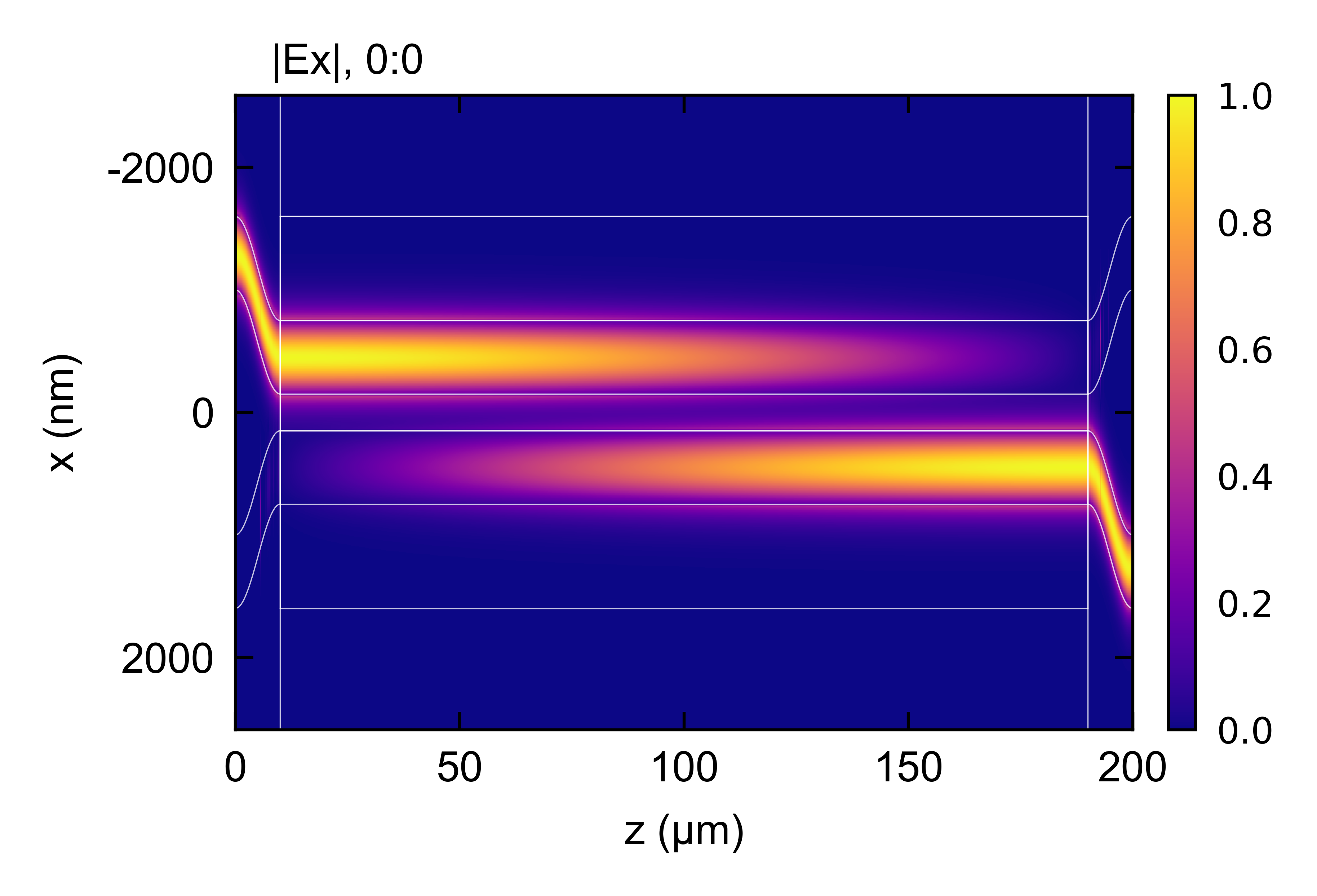
## Plot final design for 100 % coupler
em.section(name = 'coupler', length = 180e3)
em.EME()
em.plot()
## Close EMode
em.close()
%% Set simulation parameters
wavelength = 1550; % [nm] wavelength
dx = 20; dy = 20; % [nm] resolution
h_core = 220; % [nm] waveguide core height
h_clad = 1200; % [nm] waveguide top and bottom clad
width_wg = 600; % [nm]
gap = 300; % [nm]
input_gap = 2000; % [nm]
window_height = h_core + h_clad*2;
num_modes = 2; % [-] number of modes
BC = 'TE-v';
%% Connect and initialize EMode
em = emodeconnection();
%% Settings
em.settings( ...
wavelength = wavelength, x_resolution = dx, y_resolution = dy, ...
window_height = window_height, num_modes = num_modes, ...
boundary_condition = BC, background_material = 'SiO2');
%% Draw shapes
em.shape(name = 'BOX', material = 'SiO2', ...
height = h_clad);
em.shape(name = 'core', material = 'Si', ...
height = h_core, etch_depth = h_core);
%% Create profiles
em.settings(window_width = 6200);
em.label_profile(name = 'slab');
em.settings(window_width = 2600);
em.shape(name = 'core', mask = width_wg);
em.label_profile(name = 'io_waveguide');
em.settings(window_width = 2600);
em.shape(name = 'core', mask = [width_wg, width_wg], ...
mask_offset = [-gap/2-width_wg/2, gap/2+width_wg/2]);
em.label_profile(name = 'coupler');
%% Draw EME sections
em.section(name = 'input', section_type = 'straight', ...
profile = 'io_waveguide', length = 1000, ...
offset = input_gap/2 + width_wg/2);
em.section(name = 'sbend', section_type = 'gds', ...
gds = 'sbend.gds', profile = 'slab', ...
shape_to_mask = 'core', ...
length = 10e3); % modify length of gds
em.section(name = 'coupler', section_type = 'straight', ...
profile = 'coupler', length = 30e3);
em.section(section_type = 'copy', ...
section_name = 'sbend', mirror = true);
em.section(name = 'output', section_type = 'straight', ...
profile = 'io_waveguide', length = 1000, ...
offset = -input_gap/2 - width_wg/2);
%% Run EME and plot results
em.EME();
em.plot();
%% Sweep the coupler length and plot results
data = em.sweep(key = 'section, coupler, length', ...
values = 0:10000:200e3, ...
result = {'S_matrix'});
S = data.S_matrix;
figure;
plot(double(data.values)*1e-3, abs(squeeze(S(1,1,2,1,:))).^2*100);
hold on;
xlabel(append('Coupler length (', char(181), 'm)'));
ylabel('Transmission (%)');
xlim([min(data.values*1e-3) max(data.values*1e-3)]);
ylim([0 100]);
%% Plot final design for 100 % coupler
em.section(name = 'coupler', length = 180e3);
em.EME();
em.plot();
%% Close EMode
em.close();
Console output:
EMode 0.2.4 - email
Meshing completed in 0.3 sec
Meshing completed in 0.1 sec
Meshing completed in 0.1 sec
Solving S-matrices...
Meshing completed in 0.1 sec
completed in 1.5 sec
Solving section: sbend...
Solving slice at 0.0 nm... completed in 0.7 sec
Solving slice at 10000.0 nm... completed in 0.6 sec
Solving slice at 5000.0 nm... completed in 0.6 sec
Solving slice at 2500.0 nm... completed in 0.6 sec
Solving slice at 1250.0 nm... completed in 0.6 sec
Solving slice at 1875.0 nm... completed in 0.6 sec
Solving slice at 1562.5 nm... completed in 0.7 sec
Solving slice at 2187.5 nm... completed in 0.6 sec
Solving slice at 3750.0 nm... completed in 0.7 sec
Solving slice at 3125.0 nm... completed in 0.7 sec
Solving slice at 2812.5 nm... completed in 0.6 sec
Solving slice at 3437.5 nm... completed in 0.6 sec
Solving slice at 4375.0 nm... completed in 0.6 sec
Solving slice at 4062.5 nm... completed in 0.6 sec
Solving slice at 4687.5 nm... completed in 0.6 sec
Solving slice at 7500.0 nm... completed in 0.6 sec
Solving slice at 6250.0 nm... completed in 0.6 sec
Solving slice at 5625.0 nm... completed in 0.7 sec
Solving slice at 5312.5 nm... completed in 0.7 sec
Solving slice at 5937.5 nm... completed in 0.7 sec
Solving slice at 6875.0 nm... completed in 0.6 sec
Solving slice at 6562.5 nm... completed in 0.7 sec
Solving slice at 7187.5 nm... completed in 0.6 sec
Solving slice at 8750.0 nm... completed in 0.7 sec
Solving slice at 8125.0 nm... completed in 0.6 sec
Solving slice at 7812.5 nm... completed in 0.7 sec
Solving slice at 8437.5 nm... completed in 0.6 sec
Meshing completed in 0.1 sec
completed in 29.7 sec
Solving section: coupler... completed in 0.2 sec
Solving section: 3...
completed in 4.4 sec
Solving section: output... completed in 0.0 sec
completed in 0.0 sec
Meshing completed in 0.1 sec
Sweeping section parameter 'length'...
Solving EME: length = 200000.0... completed in 0.8 sec
Solving EME: length = 190000.0... completed in 0.8 sec
Solving EME: length = 180000.0... completed in 0.8 sec
Solving EME: length = 170000.0... completed in 0.8 sec
Solving EME: length = 160000.0... completed in 0.8 sec
Solving EME: length = 150000.0... completed in 0.8 sec
Solving EME: length = 140000.0... completed in 0.8 sec
Solving EME: length = 130000.0... completed in 0.8 sec
Solving EME: length = 120000.0... completed in 0.8 sec
Solving EME: length = 110000.0... completed in 0.8 sec
Solving EME: length = 100000.0... completed in 0.8 sec
Solving EME: length = 90000.0... completed in 0.8 sec
Solving EME: length = 80000.0... completed in 0.8 sec
Solving EME: length = 70000.0... completed in 0.8 sec
Solving EME: length = 60000.0... completed in 0.8 sec
Solving EME: length = 50000.0... completed in 0.8 sec
Solving EME: length = 40000.0... completed in 0.8 sec
Solving EME: length = 30000.0... completed in 0.8 sec
Solving EME: length = 20000.0... completed in 0.9 sec
Solving EME: length = 10000.0... completed in 0.8 sec
Solving EME: length = 0.0... completed in 0.8 sec
completed in 17.0 sec
Solving S-matrices...
Solving section: input... completed in 0.2 sec
Solving section: sbend... completed in 0.2 sec
Solving section: coupler... completed in 0.2 sec
Solving section: 3... completed in 0.2 sec
Solving section: output... completed in 0.0 sec
completed in 0.0 sec
Exited EMode
Figures: